![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/4-Table3-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar

Functional protein dynamics on uncharted time scales detected by nanoparticle-assisted NMR spin relaxation | Science Advances
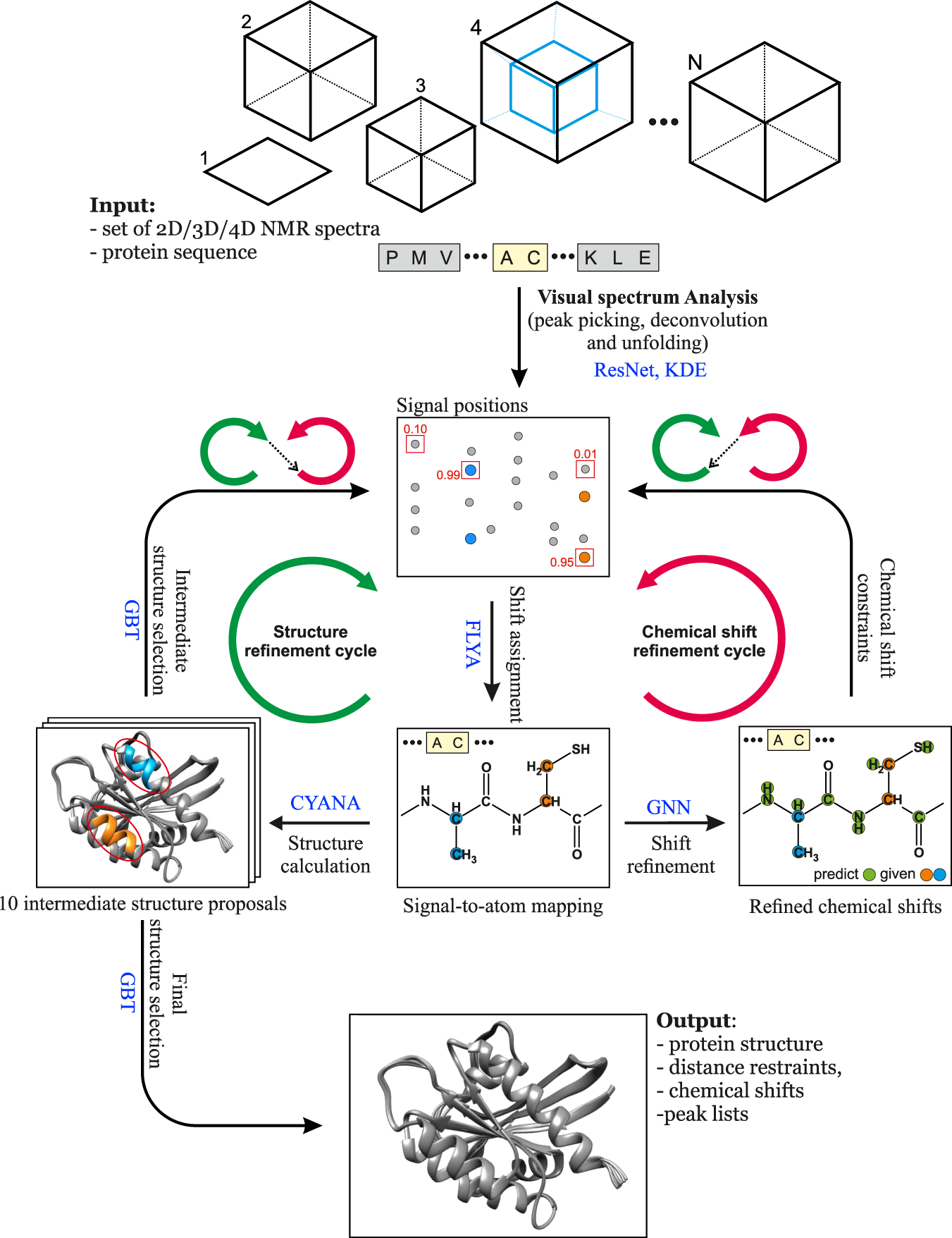
Rapid protein assignments and structures from raw NMR spectra with the deep learning technique ARTINA | Nature Communications
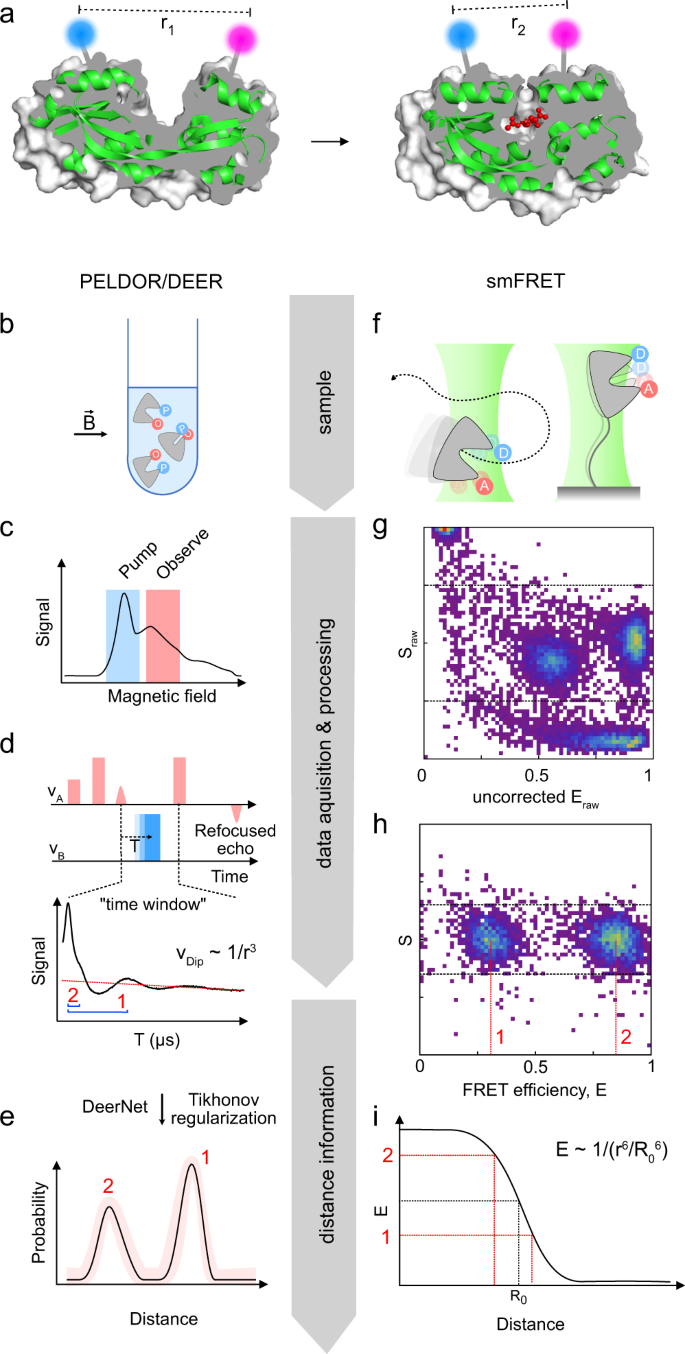
Cross-validation of distance measurements in proteins by PELDOR/DEER and single-molecule FRET | Nature Communications

Atomic resolution protein allostery from the multi-state structure of a PDZ domain | Nature Communications

A large data set comparison of protein structures determined by crystallography and NMR: Statistical test for structural differences and the effect of crystal packing - Andrec - 2007 - Proteins: Structure, Function,

Molecules | Free Full-Text | Patterns in Protein Flexibility: A Comparison of NMR “Ensembles”, MD Trajectories, and Crystallographic B-Factors

Table 1 from Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar

PDBcor: An automated correlation extraction calculator for multi-state protein structures - ScienceDirect

Chimeric single α-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology - ScienceDirect

Computational prediction of protein–protein binding affinities - Siebenmorgen - 2020 - WIREs Computational Molecular Science - Wiley Online Library
![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/3-Table2-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar

Quantitative analysis of sterol-modulated monomer–dimer equilibrium of the β1-adrenergic receptor by DEER spectroscopy | PNAS

Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with α-actinin | Science Advances
Determination of three-dimensional structures of proteins in solution by nuclear magnetic resonance spectroscopy